The Four Pillars of Understanding the Microbiome
The complex and dynamic nature of gut microbiota raises significant challenges for understanding its role in human health and diseases. To overcome these challenges, our scientific co-founder, Dr. Liping Zhao, has spent the past three decades to develop a conceptual framework encompassing Four Pillars to guide microbiome research and promote understanding of the role of gut microbiome in human health and diseases.
Pillar 1: We must move from association to causation.
To date, plenty of studies have shown that patients of various diseases have different microbiota compared to healthy individuals. There is also significant evidence identifying diet as the main force for shaping the composition of the gut microbiota. Thus, the key to changing the gut microbiota lies in dietary changes. However, in many cases it is still unclear whether changes in a patient’s gut microbiota are the cause or the end result of a disease? Changing one’s diet can simultaneously impact gut microbiota composition and patient health outcome. However, does the changed gut microbiota also improve health? Or is it merely an end result of the improved health?

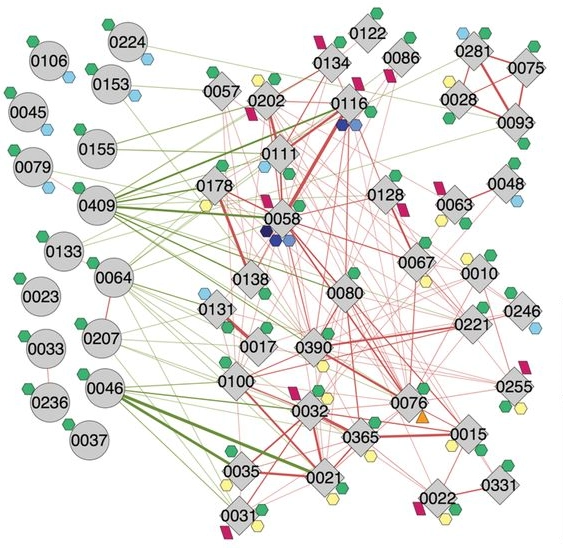
Some bacteria are more frequently found in healthy people, while other bacteria are more frequently found in patients. Nevertheless, such association evidence cannot prove whether a bacterium is beneficial or harmful. Moving from studying association to studying causality, we must isolate the gut bacteria and inoculate them into germ-free animal models to test their health effects. In 2012, Dr. Zhao found a particular pathogen overgrown in a morbidly obese patient's gut. After receiving a microbiome-targeted diet, the pathogen quickly reduced from 1/3 of the patient’s gut bacteria population to a non-detectable level. At the same time, this patient lost 110 lbs of his initial body weight and recovered from type 2 diabetes. Dr. Zhao hypothesized that this pathogen played a causal role in the patient's obesity and type 2 diabetes. This pathogen was then isolated from the patient's baseline samples and transferred to a germ-free mice gut. The lab results clearly showed that this pathogen induced obesity, fatty liver, and early diabetes in the recipient mice. Dr. Zhao then identified the exact molecule from the pathogenic bacteria that is responsible for causing the diseases. For the first time, science proved that certain “bad” gut bacteria can CAUSE obesity and type 2 diabetes.
In 2018, Dr. Zhao found that some bacteria are scarce in the gut of type 2 diabetes patients. After patients received a microbiome-targeted diet, these bacteria became abundant in the gut. Meanwhile, patients showed significant improvement in their health indicators. He then isolated these bacteria and transferred them to diabetic mice. Amazingly, the bacteria helped mice recover from their diabetes. These bacteria are proven beneficial!
We believe that conducting causality studies with a molecular level of rigor is key to establish our confidence to use bacteria and their effector molecules as biomarkers and targets for disease prevention or treatment.
Pillar 2: We must identify “good” and “bad” bacteria at the strain level.
Strains are the basic units of bacteria just as human individuals are the basic units of the human species.
As a species, human individuals share 99.9% of their genetic code which means that they are biologically similar to each other. However, bacterial strains in the same species only share 70% of their genetic code. As a reference, humans and mice still share 90% of their genetic code. This means that the difference between two strains within the same bacterial species can be greater than the difference between humans and mice.
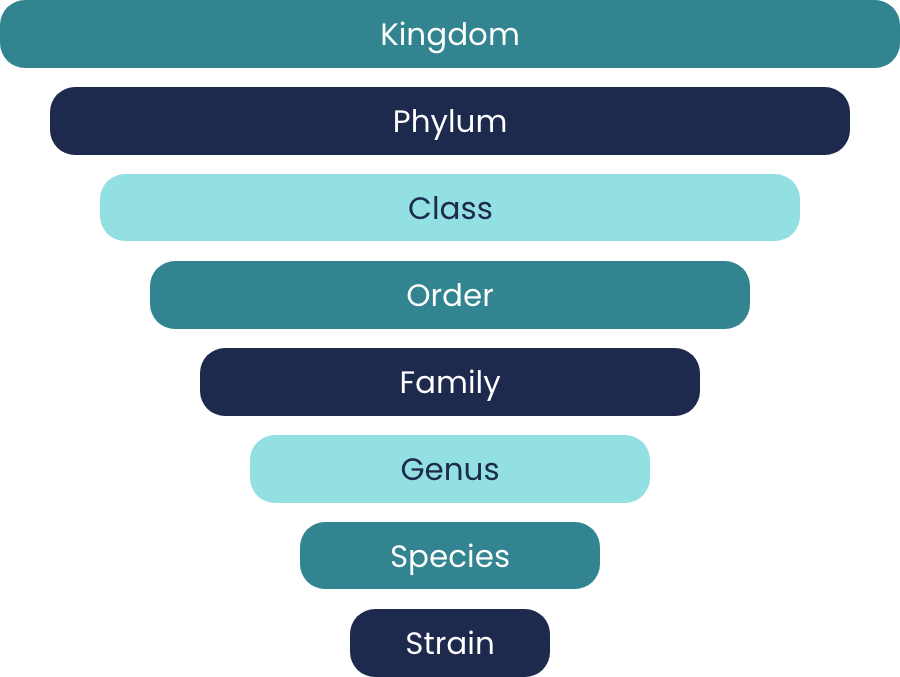
Because of such vast genetic differences within the same bacterial species, some bacteria can be pathogenic and others can be beneficial. We cannot collectively talk about bacterial species as good or bad. In 2016, Dr. Zhao found one strain that can aggravate gut inflammation and colitis in a supposedly beneficial species of gut bacteria. In 2017, he discovered that a healthy diet does not promote all the strains belonging to a supposedly beneficial species. Many of the strains were, in fact, non-responders to the healthy diet.
We cannot use species as functional units to understand the role of the gut microbiota in human health and diseases because their members often do not behave in the same way. We need to reach strain-level specificity if we desire to identify whether a bacterium is good or bad to our health.
Pillar 4: We must not rely on existing reference databases for microbiome research.
Our current knowledge of the gut microbiome is still limited. If we relied on existing reference
databases to analyze microbiome data, we would limit research to what is already known, re-discover what
others have previously found, and potentially over look novel bacteria that are important to human
health.
We need to adopt a system-level, non-targeted, discovery approach for understanding
gut microbiota in
human health and diseases. We need to develop a reference-independent strategy for analyzing
microbiome
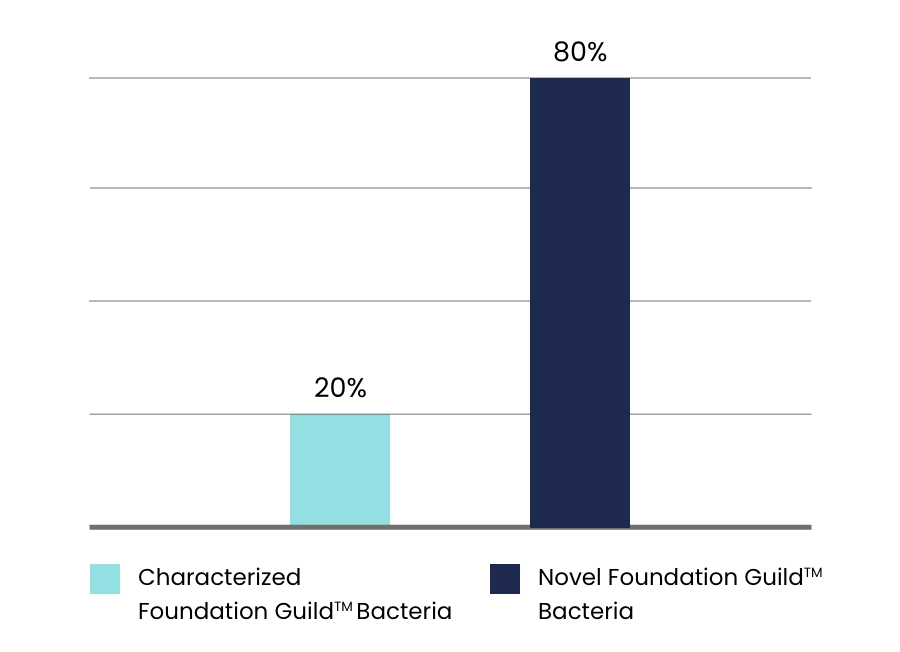
data. In the potentially beneficial guilds identified by Dr. Zhao, only about 20% can be characterized.
The other 80% represent novel bacteria that have never been discovered before.
Refrences
- Zhao, L., et al., Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science, 2018. 359(6380): p. 1151-1156.
- Costea, P.I., et al., Towards standards for human fecal sample processing in metagenomic studies. Nature biotechnology, 2017. 35(11): p. 1069.
- Zhang, X., et al., Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Scientific reports, 2015. 5: p. 14405.
- Xu, J., et al., Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. The ISME journal, 2015. 9(3): p. 552-562.
- Wang, J., et al., Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. The ISME journal, 2015. 9(1): p. 1.
- Xiao, S., et al., A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome. FEMS microbiology ecology, 2014. 87(2): p. 357-367.
- Zhao, L., The gut microbiota and obesity: from correlation to causality. Nature Reviews Microbiology, 2013. 11(9): p. 639-647.
- Zhang, C., et al., Structural modulation of gut microbiota in life-long calorie-restricted mice. Nature communications, 2013. 4(1): p. 1-10.
- Shen, J., M.S. Obin, and L. Zhao, The gut microbiota, obesity and insulin resistance. Molecular aspects of medicine, 2013. 34(1): p. 39-58.
- Fei, N. and L. Zhao, An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. The ISME journal, 2013. 7(4): p. 880-884.
- Zhang, X., et al., Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PloS one, 2012. 7(8).
- Zhang, C., et al., Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. The ISME journal, 2012. 6(10): p. 1848-1857.
- Wang, T., et al., Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. The ISME journal, 2012. 6(2): p. 320-329.
- Zhang, C., et al., Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. The ISME journal, 2010. 4(2): p. 232-241.
- Li, M., et al., Symbiotic gut microbes modulate human metabolic phenotypes. Proceedings of the National Academy of Sciences, 2008. 105(6): p. 2117-2122.